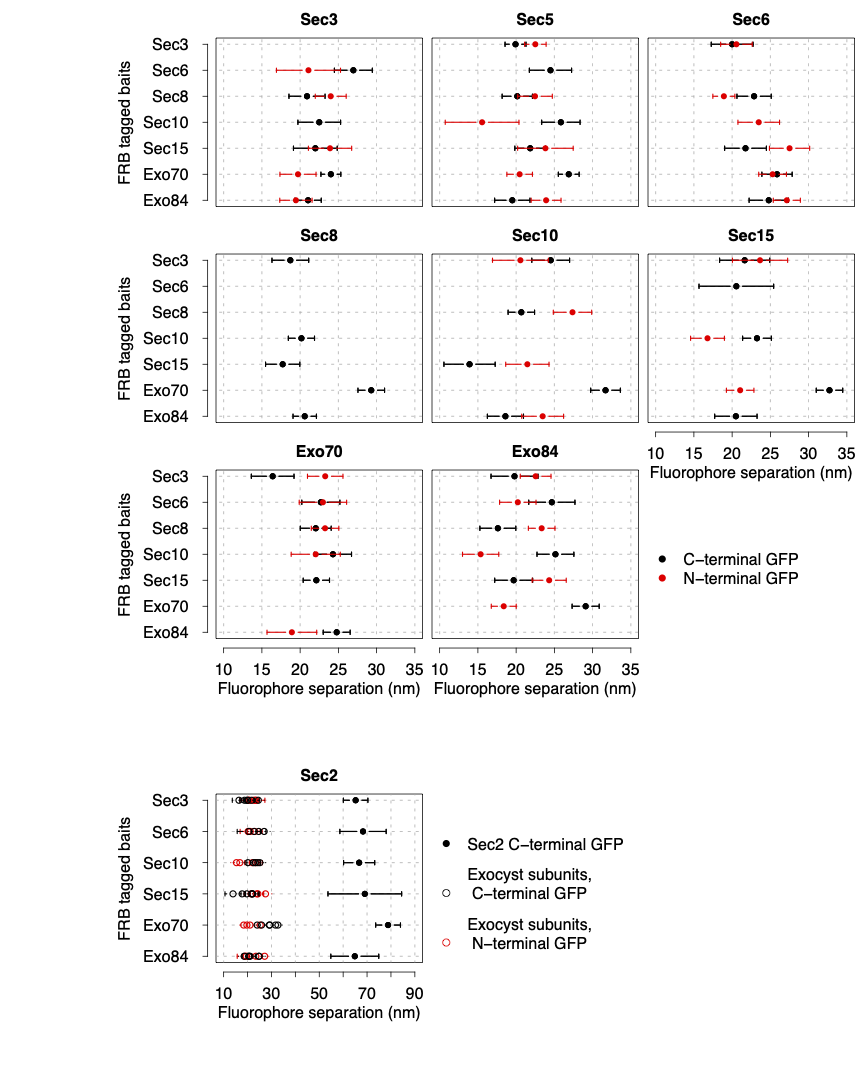
Architecture of the exocyst complex
The exocyst is a hetero-octameric complex spanning a few tens of nm. Resolving its protein organisation in living cells would be behind the optical resolution of fluorescence microscopes. We designed an innovative quantitative microscopy approach to trilaterate its architecture with unprecedented precision.
To determine its architecture, we selectively recruited exocyst complexes to an anchor tagged with mCherry. We tagged each N- and C-termini of the complex subunits one at a time with GFP, and we used the fluorophore signals to map the distance of the GFP centroid to the mCherry centroid.
We succeeded to measure the distances between the fluorophores tagging the protein termini and the anchor with nm precision. These distances allowed us to trilaterate the overall complex organisation, showing for the first time what the protein complex looks like in living cells. Our model allowed us to propose a tethering mechanism of the secretory vesicle to the plasma membrane.
References: