Tracking of endocytic dynamics
Endocytosis is a very dynamic process where a choreography of hundreds of proteins shapes the cell plasma membrane into an invagination from which a vesicle is released.
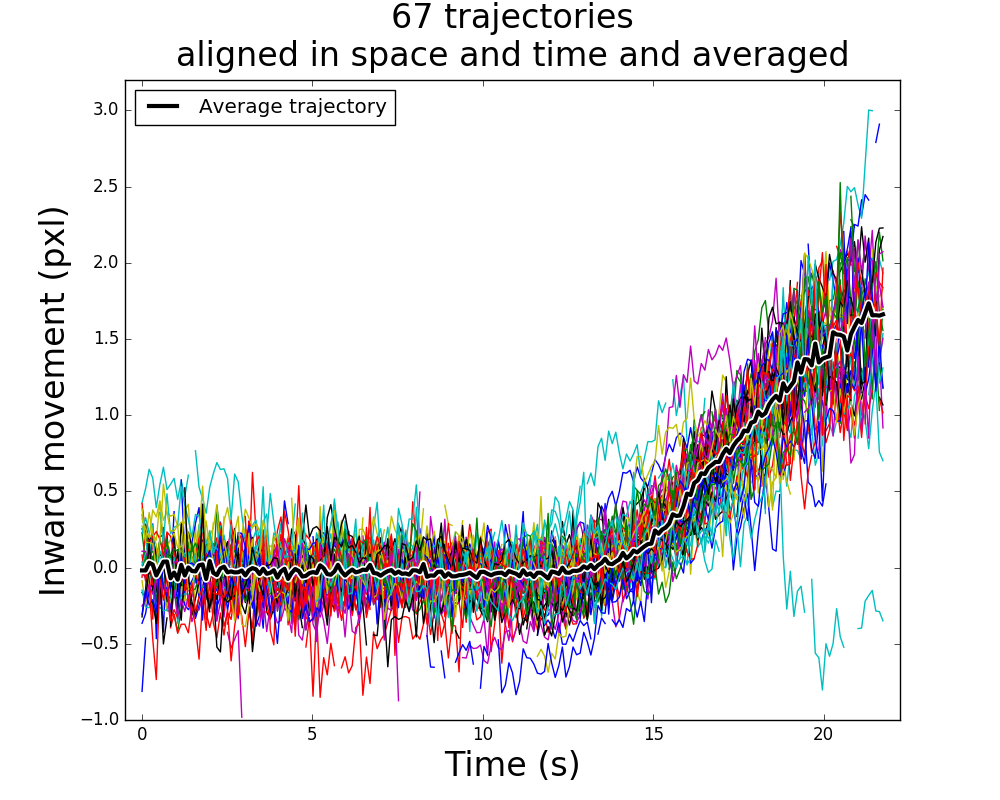
Wide-field microscopy images show endocytic events as diffraction-limited spots. The movement of these spots can be easily tracked, but it is too noisy to reliably measure the dynamics of the components of the endocytic machinery. I developed an imaging and data analysis pipeline to use these trajectories to resolve their position and dynamic properties with nm precision.
This method was later used in several publications to characterise the details of the endocytic process, mutations in its machinery, and differences between species.
References:
- Download and documentation
- Github repository
- Picco, Toret, Rivier, and Kaksonen, Biorxiv, 2024
- Manenschijn, et al., elife, 2019
- Picco, Kukulski, et al., MBoC, 2018
- Kukulski, Picco, et al., elife, 2016
- Picco, et al., elife, 2015